Data Files¶
DTDataFile¶
This is the primary interface for communication between DataTank and a Python-based helper program, assuming you use files for communication. It allows you to read, write, and append to a DataTank binary file (extension .dtbin).
-
class
datatank_py.DTDataFile.DTDataFile(file_path, truncate=False, readonly=False)¶ This class roughly corresponds to the C++ DTDataFile class.
Higher-level access is provided for some objects (e.g., PIL image), but it is primarily for writing arrays and strings. In fact, all DataTank stores is arrays and strings, with naming conventions to define how they’re interpreted.
You should not have multiple DTDataFile instances open for the same file on disk, or your file’s state will get trashed.
Reading values is fairly easy, and DTDataFile provides a dictionary-style interface to the variables. For example, assuming that a variable named “Array_One” exists:
>>> f = DTDataFile("a.dtbin") >>> v = f["Array_One"]
Setting is similar, but you’re at the mercy of the type conversion as in the write() method, and you can’t specify a time:
>>> import numpy as np >>> f = DTDataFile("a.dtbin") >>> f["My array"] = np.zeros((2, 2)) >>> f["My array"] array([[ 0., 0.], [ 0., 0.]])
Note that if you try to set the same variable name again, an exception will be thrown. Don’t do that.
You can also iterate a DTDataFile directly, and each iteration returns a variable name. Variable names are unordered, as in hashing collections.
>>> f = DTDataFile("a.dtbin") >>> for name in f: ... print name >>> if "Var" in f: ... print f["Var"]
DTDataFile supports the with statement in Python 2.5 and 2.6, so you can use this idiom to ensure resources are cleaned up:
>>> with DTDataFile("foo.dtbin", truncate=True) as df: ... df.write_2dmesh_one(mesh, 0, 0, dx, dy, "FooBar")
-
__init__(file_path, truncate=False, readonly=False)¶ Parameters: - file_path – absolute or relative path
- truncate – whether to truncate the file if it exists (default is False)
- readonly – open the file for read-only access (default is False)
The default mode is to append to a file, creating it if it doesn’t already exist. Passing True for truncate will entirely clear the file’s content.
-
close()¶ Close the underlying file object.
Further access to variables and names is not possible at this point and will raise an exception.
-
dt_object_named(key)¶ Returns: a high-level DT object, if possible, by introspection
-
ordered_variable_names()¶ Returns: list of variable names ordered as in the file
-
path()¶ Returns: the file path
-
resolve_name(name)¶ Resolve a name in case of shared variables.
Parameters: name – A potentially shared variable name Returns: The underlying variable name, with all references resolved DataTank mesh objects can be written with a shared grid, for example, and just save a string pointing to the underlying grid name. This saves a lot of disk space, but means that you can end up with a string instead of the object you’re expecting.
This method is pretty efficient in the common case of no redirect, as it only reads the header. Other cases are a bit more expensive, but too tricky to be worth rewriting at the moment.
Example from
datatank_py.DTStructuredGrid2D.DTStructuredGrid2D:name = datafile.resolve_name(name) gridx = datafile[name + "_X"] gridy = datafile[name + "_Y"] mask = DTMask.from_data_file(datafile, name + "_dom") return DTStructuredGrid2D(gridx, gridy, mask=mask)
For an example of writing a shared grid see
datatank_py.DTTriangularMesh2D.DTTriangularMesh2D.write_with_shared_grid()
-
variable_named(name, use_modules=False)¶ Procedural API for getting a value from disk.
Parameters: - name – the variable name as user-visible in the file (without the trailing nul)
- use_modules – try to convert to abstract type by introspection of available modules
Returns: a string, scalar, or numpy array
This returns values as strings, scalars, or numpy arrays. By default, no attempt is made to convert a given array to its abstract type (so you can retrieve each plane of a 2D Bitmap object by name, but not as a PIL image).
-
variable_names()¶ Returns: unsorted list of variable names
-
write(obj, name, dt_type=None, time=None)¶ Write a single value to a file object by name.
Parameters: - obj – string, numpy array, list, tuple, or scalar value
- name – user-visible name of the variable
- dt_type – string type used by DataTank
- time – time value if this variable is time-varying
Handles various object types, and adds appropriate names so they’re visible in DataTank. String, scalar, ndarray, tuple, and list objects are supported, although ndarray gives the most specific interface for precision and avoids type conversions.
This method saves a 0D array (scalar) as a “Real Number”, a 1D array as a “List of Numbers” and other shapes as “Array” by default. Use the dt_type parameter if you want something specific, such as “2D Point” for a point (although the caller has to ensure the shape is correct).
In addition, any object that implements
__dt_type__()and__dt_write__()methods can be passed, which allows saving compound types such as 2D Mesh or 2D Bitmap, without bloating up DTDataFile with all of those types.The
__dt_type__()method must return a DataTank type name:def __dt_type__(self): return "2D Mesh"
The
__dt_write__()method should use write_anonymous to save all variables as required for the object. The datafile argument is this DTDataFile instance. Note that__dt_write__()must not expose the variable by adding a “Seq” name, as that is the responsibility of DTDataFile as the higher-level object.:def __dt_write__(self, datafile, name): ... datafile.write_anonymous( ... , name)
-
write_anonymous(obj, name)¶ Write an object that will not be visible in DataTank.
Parameters: - obj – a string, numpy array, list, or tuple
- name – name of the variable
This is used for writing additional arrays and strings used by compound types, such as a 2D Mesh, which has an additional grid array.
-
write_array(array, name, dt_type=None, time=None)¶ Write an array with optional time dependence.
Parameters: - array – a numpy array, list, or tuple
- name – user-visible name of the array variable
- dt_type – string type used by DataTank
- time – time value if this variable is time-varying
This will add a string to expose it in DataTank using the dt_type parameter, which is a DataTank type such as “Array” or “NumberList.” The time parameter is a double-precision floating point value, relative to DataTank’s time slider.
Note that if time dependence is used, the caller is responsible for appending “_N” to the variable, where N is an integer >= 0 and strictly increasing with time. A contrived example follows:
>>> import numpy as np >>> f = DTDataFile("foo.dtbin") >>> for idx in xrange(10): ... point_test = np.array(range(idx, idx + 10), np.double) ... point_test = point_test.reshape((point_test.size / 2, 2)) ... tp = "2D Point Collection" ... tm = idx * 2. ... f.write_array(point_test, "Points_%d" % (idx), dt_type=tp, time=tm)
Note that the actual variable type is “2D Point Collection,” and the caller is responsible for setting the array shape correctly. This should work for any array-based object in DataTank.
-
write_string(string, name, time=None)¶ Write a string with time dependence.
Parameters: - string – the value to save
- name – the user-visible name of the string variable
- time – time value if this variable is time-varying
If this is the first time this string has been written, this method will add a string to expose it in DataTank. The time parameter is a double-precision floating point value, relative to DataTank’s time slider.
Note that if time dependence is used, the caller is responsible for appending “_N” to the variable, where N is an integer >= 0 and strictly increasing with time. A call might look like this:
>>> import datetime >>> f = DTDataFile("foo.dtbin") >>> for idx in xrange(10): ... s = datetime.now().isoformat() ... f.write_array(s, "PointTest_%d" % (idx), time=idx * 2.)
-
DTPyWrite¶
-
class
datatank_py.DTPyWrite.DTPyWrite¶ Class documenting methods that must be implemented for DTDataFile to load complex types by name.
This is never instantiated directly.
datatank_py.DTDataFile.DTDataFilechecks to ensure that an object implements all of the required methods, but you are not required to useDTPyWriteas a base class. It’s mainly provided as a convenience and formal documentation.-
__dt_type__()¶ The variable type as required by DataTank.
Returns: variable type as a string This is a string description of the variable, which can be found in the DataTank manual PDF or in DTSource. It’s easiest to look in DTSource, since you’ll need to look there for the
__dt_write__()implementation anyway. You can find the type in theWriteOne()function for a particular class, such as:// this is taken from DTPath2D.cpp void WriteOne(DTDataStorage &output,const string &name,const DTPath2D &toWrite) { Write(output,name,toWrite); Write(output,"Seq_"+name,"2D Path"); output.Flush(); }
where the type is the string “2D Path”. In some cases, it seems that multiple type names are recognized; e.g., “StringList” is written by DataTank, but “List of Strings” is used in DTSource. Regardless, this is trivial; the
datatank_py.DTPath2D.DTPath2D.__dt_type__()method looks like this:def __dt_type__(self): return "2D Path"
-
__dt_write__(datafile, name)¶ Write all associated values to a file.
Parameters: - datafile – a
datatank_py.DTDataFile.DTDataFileinstance - name – the name of the variable as it should appear in DataTank
This method collects the necessary components of the compound object and writes them to the datafile. The name is generally used as a base for associated variable names, since only one of the components can have the “primary” name. Again, the DataTank manual PDF or DTSource must be used here as a reference (DTSource is more complete). In particular, you need to look at the
Write()function implemented in the C++ class:// this is taken from DTPath2D.cpp void Write(DTDataStorage &output,const string &name,const DTPath2D &thePath) { Write(output,name+"_bbox2D",BoundingBox(thePath)); Write(output,name,thePath.Data()); }
Here the bounding box is written as name_bbox2D; this is just a 4 element double-precision array. Next, the actual path array is saved under the name as passed in to the function. The equivalent Python implementation is:
def __dt_write__(self, datafile, name): datafile.write_anonymous(self.bounding_box(), name + "_bbox2D") datafile.write_anonymous(np.dstack((self._xvalues, self._yvalues)), name)
Note that
datatank_py.DTDataFile.DTDataFile.write_anonymous()should be used in order to avoid any variable name munging (prepending “Seq_” in order to make the variable visible in DataTank).- datafile – a
-
classmethod
from_data_file(datafile, name)¶ Instantiate a
datatank_pyhigh-level object from a file.Parameters: - datafile – a
datatank_py.DTDataFile.DTDataFileinstance - name – the name of the variable
Returns: a properly initialized instance of the calling class
This class method can be implemented to read necessary components of an object from a datafile. For example:
from datatank_py.DTPath2D import DTPath2D from datatank_py.DTDataFile import DTDataFile with DTDataFile("Input.dtbin") as df: path = DTPath2D.from_data_file(df, "My Path")
will try to create a
datatank_py.DTPath2D.DTPath2Dfrom variables named “My Path” in the given data file. In general, this is the inverse of the__dt_write__()method, but may be slightly more tricky due to naming conventions in DataTank and optional data that DataTank may or may not include.- datafile – a
-
-
datatank_py.DTPyWrite.dt_writer(obj)¶ Check to ensure conformance to dt_writer protocol.
Returns: Trueif the object implements the required methods
DTSeries¶
-
class
datatank_py.DTSeries.DTSeries(datafile, series_name, series_type)¶ Base class for series support.
In general, you shouldn’t need to use this class; it’s only provided for symmetry with DTSource, and to be used by DTSeriesGroup. However, it may also be useful for non-group objects in future.
-
__init__(datafile, series_name, series_type)¶ Parameters: - datafile – an empty
datatank_py.DTDataFile.DTDataFileinstance - series_name – the name of the series variable
- series_type – the type of the series variable
The name will typically be “Var”, and the type will be whatever is the base type stored, such as “Group” for a group object.
- datafile – an empty
-
basename()¶ Returns: name of the form ‘name_N’ where N is the result of :meth:savecount
-
datafile()¶ Returns: the datatank_py.DTDataFile.DTDataFileinstance used for storage
-
last_time()¶ Returns: last time value stored or Noneif no values are stored
-
savecount()¶ Returns: the number of time values stored
Parameters: time – time value to store to disk Saves the current time value and an appropriate variable name to disk.
-
time_values()¶ Returns: vector of time values stored
-
DTSeriesGroup¶
This class allows custom objects to be created and returned, and is particularly useful when you want to compute a full time series of variables all at once.
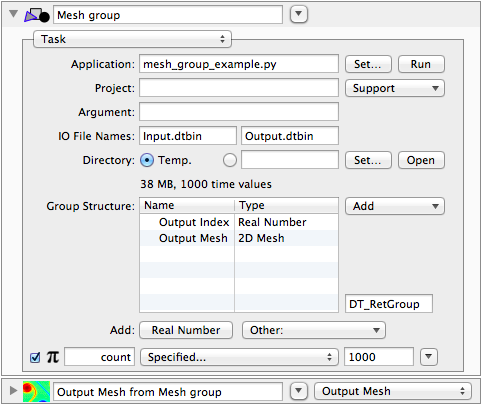
#!/usr/bin/env python
# -*- coding: utf-8 -*-
from datatank_py.DTDataFile import DTDataFile
from datatank_py.DTProgress import DTProgress
from datatank_py.DTSeries import DTSeriesGroup
from datatank_py.DTMesh2D import DTMesh2D
from time import time
import numpy as np
if __name__ == '__main__':
# dummy input file, just as boilerplate
input_file = DTDataFile("Input.dtbin")
COUNT = int(input_file["count"])
input_file.close()
start_time = time()
with DTDataFile("Output.dtbin", truncate=True) as df:
# Task groups use DTProgress for the progress bar
progress = DTProgress()
# Define the group structure using a dictionary, using the variable name as key,
# and the DataTank type as the value. This is used to create the file header.
name_to_type = { "Output Mesh":"2D Mesh", "Output Index":"Real Number" }
# Create a new DTSeriesGroup instance using that type mapping. For a task group
# to be run in DT, we want to use the "Var" name.
group = DTSeriesGroup(df, "Var", name_to_type)
# return the step to avoid getting fouled up in computing it
(x, dx) = np.linspace(-10, 10, 50, retstep=True)
(y, dy) = np.linspace(-10, 10, 100, retstep=True)
grid = (np.min(x), np.min(y), dx, dy)
xx, yy = np.meshgrid(x, y)
def mesh_function(x, y, p):
return np.cos(x + p) + np.cos(y + p)
for idx in xrange(COUNT):
mesh = mesh_function(xx, yy, idx / 10.)
group.add(idx / 10., { "Output Mesh":DTMesh2D(mesh, grid=grid), "Output Index":idx })
progress.update_percentage(float(idx) / COUNT)
# save execution time, and errors as a string list
df.write_anonymous([""], "ExecutionErrors")
df.write_anonymous(time() - start_time, "ExecutionTime")
-
class
datatank_py.DTSeries.DTSeriesGroup(datafile, name, name_to_type)¶ Base series group class.
-
__init__(datafile, name, name_to_type)¶ Parameters: - datafile – an empty
datatank_py.DTDataFile.DTDataFileinstance - name – the name of the group
- name_to_type – a dictionary mapping variable names to DataTank types
This
name_to_typedictionary defines the structure of the group:{ "My Output Array":"Array", "My Scalar Value":"Real Number" }
You can look up the DataTank type names in its PDF help manual, or for compound objects supported in
datatank_py, you can use something like:from datatank_py.DTMesh2D import DTMesh2D from datatank_py.DTPointCollection2D import DTPointCollection2D { "My 2D Mesh":DTMesh2D.dt_type[0], "My Points":DTPointCollection2D.dt_type[0] }
- datafile – an empty
-
add(time, values)¶ Add a dictionary of values.
Parameters: - time – the time value represented by these values
- values – dictionary mapping variable name to value
When adding to the group, all variables must be present, or an exception will be raised. The caller is responsible for ensuring that value types must be consistent with the expected data. Compound types (e.g., 2D Mesh) are supported via wrapper objects that implement the dt_write protocol. See DTDataFile documentation for more details.
Example:
group.add(idx / 10., { "Output Mesh":DTMesh2D(mesh, grid=grid), "Output Index":idx })
-
DTError¶
-
datatank_py.DTError.dt_set_log_identifier(ctxt)¶ Sets the default logging identifier to something useful.
Parameters: ctxt – a string that will usefully identify this log message Call this before using
DTErrorMessage()or other mechanisms; just pass in the basename of your Python script. Since DataTank changes executable names in modules (and perhaps other times), the default ofsys.argv[0]isn’t effective in tracking down error messages, as they’re all attributed to “runme” or something similar.
-
datatank_py.DTError.dt_use_syslog(should_use)¶ Allows you to copy all messages to syslog.
Parameters: should_use – pass Trueto use syslogThis can be useful in case your program croaks before results get handed back to DataTank. Currently disabled by default.
-
datatank_py.DTError.DTErrorMessage(fcn, msg)¶ Accumulate a message and echo to standard error.
Parameters: - fcn – typically a function or module name (pass
Noneto use argv[0]) - msg – an error or warning message
Typically you call this each time an error or warning should be presented, then call
DTSaveError()before exiting. This is aimed exclusively at DataTank module/external program usage.- fcn – typically a function or module name (pass
-
datatank_py.DTError.DTSaveError(datafile, name='ExecutionErrors')¶ Save accumulated messages to a file.
Parameters: - datafile – a DTDataFile instance, open for writing
- name – defaults to “ExecutionErrors” for DataTank
This will cause all messages accumulated with
DTErrorMessage()to be displayed in DataTank’s Messages panel. This is aimed exclusively at DataTank module/external program usage.
-
datatank_py.DTError.DTWarningMessage(fcn, msg)¶ Calls
DTErrorMessage(), which is what C++ DTSource does.
DTProgress¶
-
class
datatank_py.DTProgress.DTProgress¶ Drive progress indicator for DataTank.
Call update_percentage periodically to get correct progress bar timing during a long-running external program. The implementation creates a file called DTProgress in the current working directory, and DataTank reads that file to update its progress bar.
http://www.visualdatatools.com/phpBB2/viewtopic.php?t=158
-
update_percentage(percent)¶ Updates the progress indicator if needed.
Parameters: percent – percentage as floating point value (0 ≤ percent ≤ 1.0) Only updates the progress file on integral percentage points, so can be called as frequently as needed.
-